Columbia University
Irving Medical Center
Neurological Institute
710 West 168th Street, 3rd floor
(212) 305-1818
Featured Research
IN THE LAB:
Lorraine N. Clark, PhD

Lorraine N. Clark, PhD
The research focus of my lab is to investigate the genetics of neurodegenerative diseases, with a focus on Parkinson's disease (PD), Lewy body dementias (LBD), and Essential tremor (ET). Many neurodegenerative diseases share several clinical, pathological and molecular characteristics. Identifying the genetic factors that contribute to the genetic architecture of a specific neurodegenerative disease enables genotype-phenotype correlations, mechanistic studies to understand underlying disease pathogenesis, and identifies genes or pathways that can be targeted for future intervention and therapeutics.
Our current research employs a variety of genetic and genomic approaches, advanced technologies, and coordinated collaboration with clinical, statistical, and genetics researchers across disciplines to investigate 1) The role of genetic variation in the Glucocerebrosidase (GBA) gene and other lysosomal storage disorder genes and risk for PD and LBD, 2) The identification of susceptibility genes in families with early onset ET, one of the most common causes of tremor, 3) The identification of gene-gene interactions in Parkinson’s disease and 4) Ashkenazi Jewish genetics and genomics.
Results from each of these projects include:
Members of the laboratory, from left to right, Dr. Xinmin Liu, Dr. Lorraine Clark, and Sergey Kisselev.
1) We have shown previously that GBA mutations increase risk for PD. To investigate whether variants in other lysosomal storage disorder genes also contribute to disease pathogenesis we performed a genetic analysis of 241 brain autopsies from the New York Brain Bank at CU which included cases with a diagnosis of LBD, Alzheimer Disease (AD), PD and controls. A biochemical analysis, including measurement of enzyme activity and lipidomic analysis in a subset of LBD brains comparing GBA mutation carriers to non-carriers was also performed. Dr. Xinmin Liu, a postdoctoral fellow in the lab, and former postdoc Dr. Nancy Parmalee were involved in this project. Statistical analysis was performed by Taub colleague Dr. Joe Lee, and the lipidomic analysis was performed in collaboration with Taub colleagues Drs. Gil di Paolo and Robin Chan. Our studies, which are ongoing, indicate that variants in the lysosomal genes GBA, SMPD1 and MCOLN1 are associated with Lewy body pathology which has important implications for therapeutic strategies.
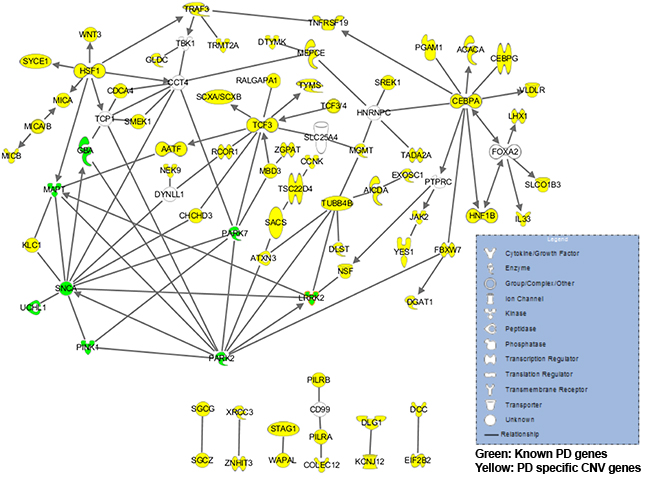 Figure 2 Ingenuity pathway analysis. Interaction of all genes included in all PD specific CNVs with known PD genes. |
2) In collaboration with Taub colleague Dr. Elan Louis, we are analyzing early onset families with ET, one of the most common causes of tremor, to identify genes that cause ET. The analysis, led by Dr Xinmin Liu, uses novel analytical methods to analyze whole exome sequence data in families that provide probabilistic predictions (genome-wide p value) for specific gene variants and genes that segregate with the ET phenotype. Other colleagues involved in this project include Drs. Ionita Laza and Ruth Ottman. Bioinformatics support was provided by Drs. Floratos and Sawle (C2B2) and sequencing was performed by Dr. Peter Nagy's lab.
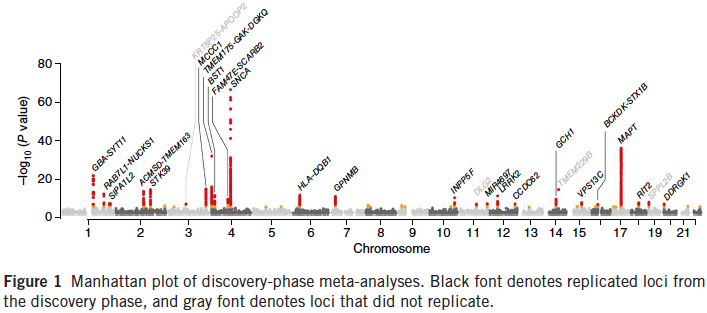
3) In a previous collaborative study with Taub colleagues Drs. Asa Abeliovich, Scott Small, and Brian McCabe, we identified a genexgene interaction between the PD genes LRRK2 and PARK16 that modifies AAO and risk for PD. Currently, we are performing a replication study and meta analysis using SNP datasets from 23andMe (a personal genomics and biotech company) and with the LRRK2 consortium led by Dr Andy Singleton (NIH) and Dr Tatiana Foroud (Indiana University).
 |
4) A particular area of interest and a rich resource for genetic studies is the Ashkenazi Jewish (AJ) population—as a genetic isolate that underwent unique population dynamics, this population is particularly conducive for powerful disease mapping. My laboratory is a part of a consortium (The Ashkenazi Genome Consortium) of investigators from the NY area with an interest in AJ genetics. The consortium led by Dr. Itsik Pe'er (Department of Computer Science, CU) has recently completed whole genome sequence and analysis of hundreds of AJ genomes. This dataset will provide a useful resource for studying complex disease in this population, as well as providing more effective personalized medicine.